In [267]:
gcn_model.eval()
out = gcn_model(data.x, data.edge_index)
visualize(out, color=data.y)
as a vector in a vector space of fixed dimension
PyG has implementations of various versions of convolution operation
The main idea is to share and filter information for neighbour nodes
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
# PyTorch geometric
! python3 -m pip install torch_geometric -q
import torch_geometric
import torch_geometric.data as geom_data
import torch_geometric.nn as geom_nn
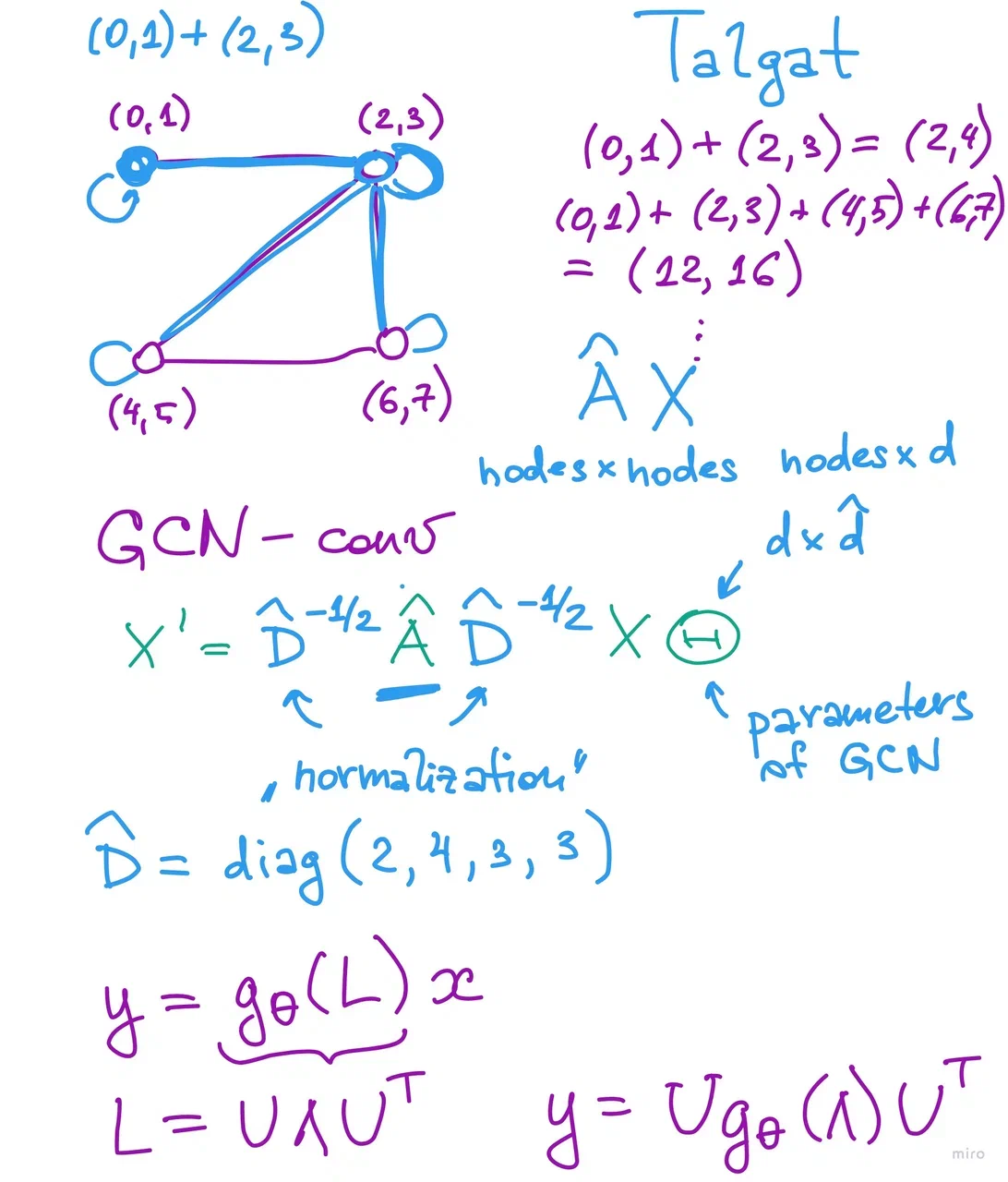
node_feats = torch.arange(8,
dtype=torch.float32).view(4, 2)
adj_matrix = torch.tensor([[1, 1, 0, 0],
[1, 1, 1, 1],
[0, 1, 1, 1],
[0, 1, 1, 1]])
print("Node features:\n", node_feats)
print("\nAdjacency matrix:\n", adj_matrix)
Node features:
tensor([[0., 1.],
[2., 3.],
[4., 5.],
[6., 7.]])
Adjacency matrix:
tensor([[1, 1, 0, 0],
[1, 1, 1, 1],
[0, 1, 1, 1],
[0, 1, 1, 1]])
# PyG takes the adjacency matrix as a list of edges
edge_list = torch.concat(torch.where(
adj_matrix == 1), axis=-1).reshape(2, -1)
print(edge_list)
tensor([[0, 0, 1, 1, 1, 1, 2, 2, 2, 3, 3, 3],
[0, 1, 0, 1, 2, 3, 1, 2, 3, 1, 2, 3]])
# Check result of simple convolution with sum aggregation
simple_conv = geom_nn.SimpleConv()
print(simple_conv(node_feats, edge_list))
tensor([[ 2., 4.],
[12., 16.],
[12., 15.],
[12., 15.]])
print(adj_matrix.float() @ node_feats)
tensor([[ 2., 4.],
[12., 16.],
[12., 15.],
[12., 15.]])
gcn = geom_nn.GCNConv(2, 8, bias=False,
normalize=True)
print(gcn(node_feats, edge_list))
-
tensor([[ 0.3723, -1.4965, -1.4036, -0.8653, 0.7523, -0.1361, -0.5825, -0.1124],
[ 0.8889, -4.9455, -4.7558, -2.3232, 2.1204, -0.8527, -2.4710, 0.0792],
[ 0.9008, -5.3986, -5.2154, -2.4269, 2.2402, -1.0128, -2.8084, 0.1782],
[ 0.9008, -5.3986, -5.2154, -2.4269, 2.2402, -1.0128, -2.8084, 0.1782]],
grad_fn=<ScatterAddBackward0>)
Parameter containing:
tensor([[-0.1523, 0.3075],
[-0.4295, -0.7643],
[-0.4919, -0.6765],
[ 0.1588, -0.6264],
[-0.0618, 0.5101],
[-0.3449, 0.0690],
[-0.5815, -0.1098],
[ 0.3099, -0.2125]], requires_grad=True)
inv_root_D = torch.diag(1. / torch.sqrt(torch.tensor([2, 4, 3, 3])))
inv_root_D @ adj_matrix.float() @ inv_root_D @ node_feats @ gcn.lin.weight.T
tensor([[ 0.3723, -1.4965, -1.4036, -0.8653, 0.7523, -0.1361, -0.5825, -0.1124],
[ 0.8889, -4.9455, -4.7558, -2.3232, 2.1204, -0.8527, -2.4710, 0.0792],
[ 0.9008, -5.3986, -5.2154, -2.4269, 2.2402, -1.0128, -2.8084, 0.1782],
[ 0.9008, -5.3986, -5.2154, -2.4269, 2.2402, -1.0128, -2.8084, 0.1782]],
grad_fn=<MmBackward0>)
gcn = geom_nn.ChebConv(2, 8, K=5)
print(gcn(node_feats, edge_list))
gcn.lins
tensor([[ 3.2364, 3.4143, -0.3946, -4.3242, -3.5987, 3.2346, -4.2101, -1.2371],
[-3.4432, -2.2127, -3.2632, -8.8110, -3.5644, 5.0098, 0.9048, 0.0408],
[-6.2562, -5.4241, -5.4977, -7.7434, 0.3508, 2.7667, 6.6166, 1.1335],
[-3.6543, -4.4635, -5.9787, -5.9360, 3.6287, 1.4175, 7.0212, -0.2499]],
grad_fn=<AddBackward0>)
ModuleList( (0): Linear(2, 8, bias=False) (1): Linear(2, 8, bias=False) (2): Linear(2, 8, bias=False) (3): Linear(2, 8, bias=False) (4): Linear(2, 8, bias=False) )
from torch_geometric.datasets import Planetoid
from torch_geometric.transforms import NormalizeFeatures
dataset = Planetoid(root='data/Planetoid', name='Cora')
print('Number of graphs: {}'.format(len(dataset)))
print('Number of features: {}'.format(dataset.num_features))
print('Number of classes: {}'.format(dataset.num_classes))
data = dataset[0] # Get the first graph object.
print(data)
print(data.x[0], data.y[0].item())
Number of graphs: 1 Number of features: 1433 Number of classes: 7 Data(x=[2708, 1433], edge_index=[2, 10556], y=[2708], train_mask=[2708], val_mask=[2708], test_mask=[2708]) tensor([0., 0., 0., ..., 0., 0., 0.]) 3
%matplotlib inline
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
def visualize(h, color):
z = TSNE(n_components=2).fit_transform(h.detach().cpu().numpy())
plt.figure(figsize=(10,10))
plt.xticks([])
plt.yticks([])
plt.scatter(z[:, 0], z[:, 1], s=70, c=color)
plt.show()
class GCN(torch.nn.Module):
def __init__(self, hidden_channels):
super().__init__()
torch.manual_seed(2128506)
self.conv1 = geom_nn.GCNConv(dataset.num_features, hidden_channels)
self.conv2 = geom_nn.GCNConv(hidden_channels, dataset.num_classes)
self.drop = torch.nn.Dropout(0.5)
def forward(self, x, edge_index):
x = self.conv1(x, edge_index)
x = x.relu()
x = self.drop(x)
x = self.conv2(x, edge_index)
return x
gcn_model = GCN(hidden_channels=32)
print(gcn_model)
GCN( (conv1): GCNConv(1433, 32) (conv2): GCNConv(32, 7) (drop): Dropout(p=0.5, inplace=False) )
gcn_model.eval()
out = gcn_model(data.x, data.edge_index)
visualize(out, color=data.y)
criterion = torch.nn.CrossEntropyLoss()
def test(model, data):
model.eval()
out = model(data.x, data.edge_index)
pred = out.argmax(dim=1)
test_correct = pred[data.test_mask] == data.y[data.test_mask]
test_acc = int(test_correct.sum()) / int(data.test_mask.sum())
return test_acc
def train(model, data, optimizer):
model.train()
optimizer.zero_grad()
out = model(data.x, data.edge_index)
loss = criterion(out[data.train_mask], data.y[data.train_mask])
loss.backward()
optimizer.step()
return loss
optimizer = torch.optim.Adam(gcn_model.parameters(), lr=0.01)
gcn_test_accuracy = []
for epoch in range(1, 21):
loss = train(gcn_model, data, optimizer)
gcn_test_accuracy.append(test(gcn_model, data))
print(f'Epoch: {epoch:},\t Loss: {loss:.4f}')
Epoch: 1, Loss: 1.9275 Epoch: 2, Loss: 1.7472 Epoch: 3, Loss: 1.5225 Epoch: 4, Loss: 1.3219 Epoch: 5, Loss: 1.1181 Epoch: 6, Loss: 0.9258 Epoch: 7, Loss: 0.7446 Epoch: 8, Loss: 0.6071 Epoch: 9, Loss: 0.4898 Epoch: 10, Loss: 0.4167 Epoch: 11, Loss: 0.3402 Epoch: 12, Loss: 0.2981 Epoch: 13, Loss: 0.2287 Epoch: 14, Loss: 0.1905 Epoch: 15, Loss: 0.1524 Epoch: 16, Loss: 0.1184 Epoch: 17, Loss: 0.1126 Epoch: 18, Loss: 0.1082 Epoch: 19, Loss: 0.0678 Epoch: 20, Loss: 0.0586
test_acc = test(gcn_model, data)
print(f'Test Accuracy: {test_acc:.4f}')
Test Accuracy: 0.7800
gcn_model.eval()
out = gcn_model(data.x, data.edge_index)
visualize(out, color=data.y)
plt.plot(gcn_test_accuracy)
[<matplotlib.lines.Line2D at 0x7fb1ff041070>]
class GChebCN(torch.nn.Module):
def __init__(self, hidden_channels, K):
super().__init__()
torch.manual_seed(2128506)
self.conv1 = geom_nn.ChebConv(dataset.num_features, hidden_channels, K)
self.conv2 = geom_nn.ChebConv(hidden_channels, dataset.num_classes, K)
self.drop = torch.nn.Dropout(0.5)
def forward(self, x, edge_index):
x = self.conv1(x, edge_index)
x = x.relu()
x = self.drop(x)
x = self.conv2(x, edge_index)
return x
cheb_gcn = GChebCN(32, K=3)
optimizer = torch.optim.Adam(cheb_gcn.parameters(), lr=0.01)
chebcn_test_accuracy = []
for epoch in range(1, 21):
loss = train(cheb_gcn, data, optimizer)
chebcn_test_accuracy.append(test(cheb_gcn, data))
print(f'Epoch: {epoch},\t Loss: {loss:.4f}')
Epoch: 1, Loss: 2.0385 Epoch: 2, Loss: 1.3648 Epoch: 3, Loss: 0.7926 Epoch: 4, Loss: 0.4739 Epoch: 5, Loss: 0.2473 Epoch: 6, Loss: 0.1637 Epoch: 7, Loss: 0.0903 Epoch: 8, Loss: 0.0440 Epoch: 9, Loss: 0.0396 Epoch: 10, Loss: 0.0246 Epoch: 11, Loss: 0.0341 Epoch: 12, Loss: 0.0168 Epoch: 13, Loss: 0.0097 Epoch: 14, Loss: 0.0062 Epoch: 15, Loss: 0.0072 Epoch: 16, Loss: 0.0213 Epoch: 17, Loss: 0.0035 Epoch: 18, Loss: 0.0043 Epoch: 19, Loss: 0.0070 Epoch: 20, Loss: 0.0008
test_acc = test(cheb_gcn, data)
print(f'Test Accuracy: {test_acc:.4f}')
Test Accuracy: 0.7750
cheb_gcn.eval()
out = cheb_gcn(data.x, data.edge_index)
visualize(out, color=data.y)
plt.plot(gcn_test_accuracy, label="GCN")
plt.plot(chebcn_test_accuracy, label="Cheb CN")
plt.legend()
plt.ylabel("Test accuracy")
plt.xlabel("# epoch")
Text(0.5, 0, '# epoch')
where $a$ is some predefined function and $W$ is learnable matrix
where $u \in \mathbb{R}^{dim(x) + dim(y)}$ is a learnable vector
class GAT(torch.nn.Module):
def __init__(self, hidden_channels, heads):
super().__init__()
torch.manual_seed(2128506)
self.conv1 = geom_nn.GATConv(dataset.num_features, hidden_channels,heads)
self.conv2 = geom_nn.GATConv(heads * hidden_channels, dataset.num_classes,heads)
self.drop = torch.nn.Dropout(0.5)
self.relu = nn.ReLU()
def forward(self, x, edge_index):
x = self.drop(x)
x = self.conv1(x, edge_index)
x = self.relu(x)
x = self.drop(x)
x = self.conv2(x, edge_index)
return x
model = GAT(hidden_channels=32, heads=8)
print(model)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
gat_test_accuracy = []
for epoch in range(1, 21):
loss = train(model, data, optimizer)
gat_test_accuracy.append(test(model, data))
print(f'Epoch: {epoch:},\t Loss: {loss:.4f}')
GAT( (conv1): GATConv(1433, 32, heads=8) (conv2): GATConv(256, 7, heads=8) (drop): Dropout(p=0.5, inplace=False) (relu): ReLU() ) Epoch: 1, Loss: 3.9761 Epoch: 2, Loss: 3.0069 Epoch: 3, Loss: 1.8436 Epoch: 4, Loss: 1.0772 Epoch: 5, Loss: 0.6553 Epoch: 6, Loss: 0.4706 Epoch: 7, Loss: 0.2994 Epoch: 8, Loss: 0.2233 Epoch: 9, Loss: 0.1750 Epoch: 10, Loss: 0.1970 Epoch: 11, Loss: 0.1165 Epoch: 12, Loss: 0.1241 Epoch: 13, Loss: 0.0932 Epoch: 14, Loss: 0.1476 Epoch: 15, Loss: 0.1312 Epoch: 16, Loss: 0.1232 Epoch: 17, Loss: 0.0600 Epoch: 18, Loss: 0.1695 Epoch: 19, Loss: 0.1520 Epoch: 20, Loss: 0.0580
test_acc = test(model, data)
print(f'Test Accuracy: {test_acc:.4f}')
Test Accuracy: 0.7200
plt.plot(gcn_test_accuracy, label="GCN")
plt.plot(chebcn_test_accuracy, label="Cheb CN")
plt.plot(gat_test_accuracy, label="Graph attention")
plt.legend()
plt.ylabel("Test accuracy")
plt.xlabel("# epoch")
Text(0.5, 0, '# epoch')
model.eval()
out = model(data.x, data.edge_index)
visualize(out, color=data.y)
where $\mathbf{x}_i$ are edge embeddings and $h_{\mathbf{\Theta}}$ is an MLP.